Modelling Targeted Protein Degraders (TPDs) Utilizing Computational And Medicinal Chemistry Approaches
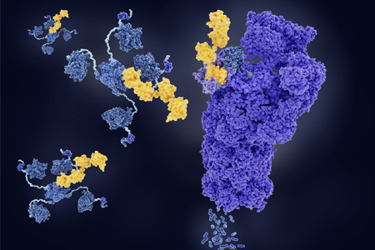
Designing and developing targeted protein degraders presents numerous complexities. The proteins selected for degradation are typically implicated in disease pathways and possess intricate, dynamic structures that make it difficult to identify suitable binding regions or vulnerabilities. Even when viable binding sites are found on both the target protein and an E3 ligase, engineering and synthesizing inhibitor ligands that can engage the ubiquitin-proteasome system effectively remains a significant hurdle. One of the most critical challenges lies in designing the linker that connects these two ligands. This component must strike an optimal balance in terms of its length, flexibility, and composition to ensure the resulting degrader molecule is not only chemically stable but also exhibits adequate solubility and cell permeability—attributes essential for intracellular activity.
This case study highlights the success of Aragen Life Sciences’ Integrated Drug Discovery (IDD) team in overcoming these obstacles. Through a carefully executed strategy, the team identified novel degrader molecules with the necessary physicochemical properties to achieve robust degradation of the target Protein of Interest (POI).
Get unlimited access to:
Enter your credentials below to log in. Not yet a member of Outsourced Pharma? Subscribe today.
