TARGATT™ Large Knock-In Technology

Co-invented by Applied StemCell’s President and Chief Scientific Officer, Ruby Tsai, Ph.D., and Head of R&D, Alfonso Farruggio, Ph.D., TARGATT™ large knock-in technology enables rapid, efficient, and precise integration of large DNA fragments—up to 20 kb in a single reaction, more in nested reactions—into a specific intergenic locus.
Built To Overcome The Challenges Of Random Integration
Because integration is precise, single copy, and targeted to a defined site, we can avoid the challenges encountered with random integration mechanisms such as position effects, gene silencing, and gene instability due to the integration of multiple transgene copies.
These same features also enable time-saving capabilities such as working with pools, whether you are comparing variants or simply need stable gene expression. While you can also do these things with systems that integrate randomly, it takes more time and effort.
Taken together, these features make TARGATT™ an incredibly versatile tool for stable mammalian cell line development.
Expand What You Can Accomplish In Mammalian Cells With TARGATT™ Technology

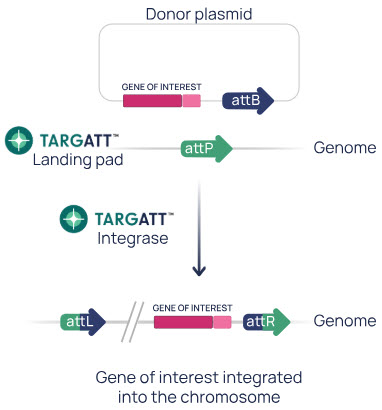
How TARGATT™ technology works
- The TARGATT™ platform consists of a serine integrase and two DNA sequences—the attB site and the attP site. The donor plasmid will contain one of these sites and your DNA payload and the genome will contain the other site as part of the TARGATT™ landing pad, which we have pre-engineered into the H11 safe harbor locus.
- We can also customize the location of the TARGATT™ landing pad for your project.
- The TARGATT™ integrase binds the attP and attB sites, mediating a site-specific recombination event that results in the stable insertion of a single copy of the donor plasmid into the genome at the attP site.
- Because the attB and attP sites are destroyed during the recombination event, the integration event is unidirectional resulting in stable and efficient integration.
- The TARGATT™ platform consists of a serine integrase and two DNA sequences—the attB site and the attP site. The donor plasmid will contain one of these sites and your DNA payload and the genome will contain the other site as part of the TARGATT™ landing pad, which we have pre-engineered into the H11 safe harbor locus.
- We can also customize the location of the TARGATT™ landing pad for your project.
- The TARGATT™ integrase binds the attP and attB sites, mediating a site-specific recombination event that results in the stable insertion of a single copy of the donor plasmid into the genome at the attP site.
- Because the attB and attP sites are destroyed during the recombination event, the integration event is unidirectional resulting in stable and efficient integration.
Why You Should Choose TARGATT™ Technology
Efficient integration, consistent expression
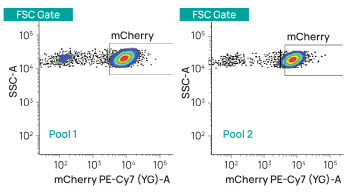
Figure 1. TARGATT™ technology delivers efficient integration with consistent expression. We inserted mCherry into the TARGATT™ landing pad at the H11 safe harbor site in HEK293 cells and used flow cytometry to assess mCherry expression in two different pools of integrants.
Our optimized system consistently delivers >90% integration efficiency after drug selection (as much as 50% before drug selection). Unlike CRISPR Cas9, TARGATT™ technology does not rely on the host’s homologous recombination machinery, enabling its use in non-dividing cells.
Expression from the H11 locus is highly consistent, as can be seen in the reproducibly tight clustering of the mCherry signal in Figure 1.
Large cargo insertion
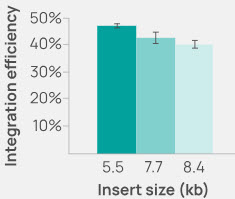
Figure 2. Raw integration efficiency (no drug selection) into HEK293 cells remains high as insert size increases from 5.5 to 8.4 kb.
Integration is highly efficient even with inserts of 20 kb. Larger insertions can be accommodated with multiple, nested integrations.
This opens the door to expressing multiple proteins while maintaining desired stoichiometric ratios.
Site-specific integration
TARGATT™ technology is highly specific for the corresponding landing pad, resulting in negligible off-target events.
For our service projects, we verify on-target insertion using Sanger sequencing at the insertion junctions, and can provide additional characterization should your project require.
Automation-ready, pool-friendly
Highly efficient integration into the robust, reliably-expressing H11 locus means you can be confident that your inserted genes will be expressed* and you don’t need to hunt for the optimal clone. These features make TARGATT™ technology compatible with streamlined pool-based studies and automated workflows.
*For constructs that have demonstrated expression in mammalian cells.
Single copy, unidirectional insertion

Figure 3. Integration using TARGATT technology is single copy. (A) We co-transfected GFP and mCherry TARGATT™ donor plasmids into CHO cells and imaged the resulting cells using the green channel (B), the red channel (C), and as an overlay (D). The lack of yellow cells in the overlay indicates the absence of cells expressing both GFP and mCherry, demonstrating that genomic insertion only occurs in single copy.
Because TARGATT™ integration occurs through a controlled, site-specific mechanism, only a single copy of the DNA insert is incorporated into the genome.
The integration process destroys the attP site in the landing pad rendering the process unidirectional and, thus, highly efficient.
What You Can Achieve With TARGATT™ Technology
TARGATT™ large knock-in technology accelerates and enables a range of cell engineering applications due to a combination of highly efficient integration and robust, reproducible expression from the H11 locus.
Engineer faster
Say goodbye to worrying about getting a good integrant and hunting for the right clone.
Experiment faster
Start studies sooner, work more efficiently when you can use pools instead of purified clones.
Do more in mammalian cells
Run studies in more relevant cell types to save time and improve your chances for success in the clinic.
Applications
- Screening
- Create screening libraries of 107 to 109 in mammalian cells
- Assess variants of full-length proteins and antibodies
- Test the effects of small molecules, proteins, antibodies on promoters
- Protein expression
- Express multiprotein complexes
- Express challenging proteins such as membrane proteins
- Express with strict stoichiometry, such as for bispecific antibodies
- Gene therapy
- Build stable packaging lines
- Cell therapy
- Knock-in CAR constructs and other proteins
- Construct multi-component CARs with logic gates
- Disease modeling
- Create strains that express your gene(s) of interest
